Methodological repertoire
About the ITC Calorimetry
Contact me to take an appointment E.Ennifar@ibmc-cnrs.unistra.fr or K.Brillet@ibmc-cnrs.unistra.fr and to book a time slot. We ask 168.77 € HT for CNRS and Strasbourg University labs or 383.56 € HT for external labs for each 4 h time slot. A typical ITC experiment requires than 1 hour.
Isothermal Titration Calorimetry (ITC) (microcal website) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method.
ITC200 is the most sensitive isothermal titration calorimeter available, but it is also extremely fragile. For that reason, I will take care of it and perform your experiment(s) (but not process your data !)
Filtrer ou centrifuger vos échantillons (très important surtout pour éviter toute obstruction de la seringue).
Samples requirement : 300 µl of sample in the cell
70 µl of ligand in the syringe (up to 200 µl if possible)
10 ml of buffer solution
Sample and ligand have to be strictly in the same buffer (salts, pH …). This is extremely important for the ITC measurement ! Any difference will generate noise and make interpretation more difficult.
To approximate the concentration needed in the cell, estimate the Kd for the system and multiply by 10. With very tight binders, the concentration should be on the low side of this value ; for weak binders, the concentration should be on the high side of this value. Multiply the cell concentration by n and by 10 – 15 for the syringe concentration. The minimum and maximum concentrations that are commonly used in the ITC200 are 3 µM to 500 µM.
ΔS and ΔG are deduced from observed Kd and ΔH : ΔG = -RT log (1/Kd) = ΔH – T ΔS The binding reaction should be either endothermic or exothermic (ΔH ≠ 0), otherwise no signal will be observed.
The binding reaction should be either endothermic or exothermic (ΔH ≠ 0), otherwise no signal will be observed.
An optimal determination of the Kd is obtained if 1000 > c > 1, where c = number of sites x Ka x concentration. For an optimal determination of the ΔH, a plateau should be obtained at the beginning of the experiment.
Process your data either with your favourite program or with Origin 7 installed on the ITC200 computer (see the manual below).
kinITC : a new method for obtaining joint thermodynamic and kinetic data by Isothermal Titration Calorimetry
D. Burnouf, E. Ennifar, S. Guedich, B. Puffer, G. Hoffmann, G. Bec, F. Disdier, M. Baltzinger & P. Dumas
JACS (2011) DOI : 10.1021/ja209057d
http://pubs.acs.org/doi/abs/10.1021/ja209057d
Performance specifications
Operating temperature range : 2 – 80°C
Titration sensitivity quotient : 8 ncal × ml
Response time : 10 s
Cell : 200 µl
Injection syringe : 40 µl
Smallest injection : 0.1 µl

The ITC200 is now capable of carrying out a complete binding experiment using only a single continuous injection as opposed to the normal procedure which requires multiple injections. In this single-injection procedure, only one slow, continuous injection of ligand solution is made from the injection syringe into the macromolecule solution contained in the sample cell. The SIM technique significantly improves the speed of the experiment (typically 6 – 12 minutes per run !) and is particularly appropriate for drug discovery and target screening. Data analysis should be performed with the Origin Microcal LLC SIM module.
Example of SIM performed on our ITC200 (19/01/2009) and comparison with an equivalent multiple injection run :
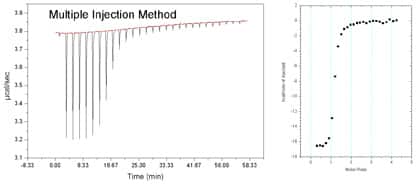

More information about the SIM is available on the Microcal website. Also have a look into documentations available on this site.
Accueil > Recherche > Équipes & Axes de Recherche > Structure et dynamique des machines biomoléculaires – E. Ennifar > ITC microcalorimetry
ITC microcalorimetry
- The use of biophysical methods increases success in obtaining liganded crystal structures
- Isothermal titration calorimetry of RNA
- Applications of Isothermal Titration Calorimetry in RNA Biochemistry and Biophysics
- Heat Capacity Changes Associated with Nucleic Acid Folding
- Calorimetry and Thermodynamics in Drug Design
- Ligand binding to one-dimensional lattice-like macromolecules : Analysis of the McGhee–von Hippel theory implemented in isothermal titration calorimetry
- ITC in the post-genomic era. . .? Priceless
- Isothermal titration calorimetry to determine association constants for high-affinity ligands
- Thermodynamics of Aminogl.–rRNA Recognition
http://pubs.acs.org/doi/abs/10.1021/ja209057d Supplementary Information
 Isothermal Titration Calorimetry (ITC) (microcal website) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method.
What do you need for an experiment ?
Contact me to take an appointment E.Ennifar@ibmc-cnrs.unistra.fr or K.Brillet@ibmc-cnrs.unistra.fr and to book a time slot. We ask 168.77 € HT for CNRS and Strasbourg University labs or 383.56 € HT for external labs for each 4 h time slot. A typical ITC experiment requires than 1 hour.
Isothermal Titration Calorimetry (ITC) (microcal website) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method.
What do you need for an experiment ?
Contact me to take an appointment E.Ennifar@ibmc-cnrs.unistra.fr or K.Brillet@ibmc-cnrs.unistra.fr and to book a time slot. We ask 168.77 € HT for CNRS and Strasbourg University labs or 383.56 € HT for external labs for each 4 h time slot. A typical ITC experiment requires than 1 hour.
ITC200 is the most sensitive isothermal titration calorimeter available, but it is also extremely fragile. For that reason, I will take care of it and perform your experiment(s) (but not process your data !) Filter or centrifuge your samples (very important especially to avoid any obstruction of the syringe) Samples requirement : 300 µl of sample in the cell 70 µl of ligand in the syringe (up to 200 µl if possible) 10 ml of buffer solution Sample and ligand have to be strictly in the same buffer (salts, pH …). This is extremely important for the ITC measurement ! Any difference will generate noise and make interpretation more difficult. To approximate the concentration needed in the cell, estimate the Kd for the system and multiply by 10. With very tight binders, the concentration should be on the low side of this value ; for weak binders, the concentration should be on the high side of this value. Multiply the cell concentration by n and by 10 – 15 for the syringe concentration. The minimum and maximum concentrations that are commonly used in the ITC200 are 3 µM to 500 µM. ΔS and ΔG are deduced from observed Kd and ΔH : ΔG = -RT log (1/Kd) = ΔH – T ΔS The binding reaction should be either endothermic or exothermic (ΔH ≠ 0), otherwise no signal will be observed An optimal determination of the Kd is obtained if 1000 > c > 1, where c = number of sites x Ka x concentration. For an optimal determination of the ΔH, a plateau should be obtained at the beginning of the experiment Process your data either with your favourite program or with Origin 7 installed on the ITC200 computer (see the manual below) Performance specifications :
- Operating temperature range : 2 – 80°C
- Titration sensitivity quotient : 8 ncal × ml
- Response time : 10 s
- Cell : 200 µl
- Injection syringe : 40 µl
- Smallest injection : 0.1 µl

TC experiment performed on the Microcal ITC200 : successive injections of 1.4 µl lividomycin aminoglycoside 250 µM in 12 µM HIV-1 DIS RNA
Single Injection Method (SIM) The ITC200 is now capable of carrying out a complete binding experiment using only a single continuous injection as opposed to the normal procedure which requires multiple injections. In this single-injection procedure, only one slow, continuous injection of ligand solution is made from the injection syringe into the macromolecule solution contained in the sample cell. The SIM technique significantly improves the speed of the experiment (typically 6 – 12 minutes per run !) and is particularly appropriate for drug discovery and target screening. Data analysis should be performed with the Origin Microcal LLC SIM module. Example of SIM performed on our ITC200 (19/01/2009) and comparison with an equivalent multiple injection run :
 Multiple 1.4 µl injections of 250 µM lividomycin aminoglycoside into a 12 µM HIV-1 DIS RNA solution.
Multiple 1.4 µl injections of 250 µM lividomycin aminoglycoside into a 12 µM HIV-1 DIS RNA solution.
 Single injection of 36 µl lividomycin 250 µM into a 12 µM HIV-1 DIS RNA solution.
More information about the SIM is available on the Microcal website (http://www.microcal.com/products/itc/single-injection-method.asp). Also have a look into documentations available on this site.
What is ITC ? (from Microcal website)
Isothermal Titration Calorimetry (ITC) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method.
Single injection of 36 µl lividomycin 250 µM into a 12 µM HIV-1 DIS RNA solution.
More information about the SIM is available on the Microcal website (http://www.microcal.com/products/itc/single-injection-method.asp). Also have a look into documentations available on this site.
What is ITC ? (from Microcal website)
Isothermal Titration Calorimetry (ITC) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method.
When substances bind, heat is either generated or absorbed. ITC is a thermodynamic technique that directly measures the heat released or absorbed during a biomolecular binding event. Measurement of this heat allows accurate determination of binding constants (KB), reaction stoichiometry (n), enthalpy (ΔH) and entropy (ΔS), thereby providing a complete thermodynamic profile of the molecular interaction in a single experiment. Because ITC goes beyond binding affinities and can elucidate the mechanism of the molecular interaction, it has become the method of choice for characterizing biomolecular interactions.
Applications include:
- Characterization of molecular interactions of small molecules, proteins, antibodies, nucleic acids, lipids and other biomolecules.
- Lead optimization.
- Enzyme kinetics.
- Assessment of the effect of molecular structure changes on binding mechanisms.
- Assessment of biological activitiy.
Binding Studies
All cellular processes require specific binding and molecular recognition between biomolecules. Knowledge of these interactions is important to understand how proteins, nucleic acids and lipids function in biological systems. There have been rapid advances in structural biology and relating structure to biochemical function and mechanism. However, knowledge of structure alone does not ensure accurate prediction of function and biological activity. The complete characterization of any binding interaction requires a quantification of the affinity, number of binding sites and the thermodynamics.
Thermodynamic data, specifically enthalpy (ΔH) and entropy (ΔS), reveal the forces that drive complex formation and mechanism of action. Thermodynamics provide information on conformational changes, hydrogen bonding, hydrophobic interactions and charge-charge interactions. This information can be used to describe the function and mechanism at a molecular level.
Binding Studies with Isothermal Titration Calorimetry (ITC)
Isothermal Titration Calorimetry (ITC) is a powerful analytical tool which measures the binding affinity and thermodynamics between any two biomolecules. ITC is considered the “gold standard” assay for binding, and has many advantages:
- Universal assay – directly measures heat change associated with binding
- Label-free – uses native materials
- True in-solution technique
- Requires minimal assay development
- Has no molecular weight limitations
- Non-optical
- Versatile, can be used with any biomolecule – proteins, nucleic acids, small molecule drugs, lipids, etc.
- Can be used with a wide range of biological buffers, ionic strengths, pH’s
In a single ITC experiment, one can determine:
- Binding affinity – Kd
- Directly measure sub-millimolar to nanomolar
- Can extend affinity range to picomolar using competitive ITC method
- Number of binding sites
- Multiple and different binding sites
- Enthalpy (ΔH) and entropy (ΔS) of binding
ITC is vital in the study of multi-probe structure activity relationships (SAR) since it can detect contributions that affinity-only methods may miss. For example, the affinity measured by these methods may be similar for a wild-type and mutant protein binding to a drug, but ITC can reveal differences in ΔH and ΔS that can describe the mechanism of action of binding. This information can validate in-silico modeling. ITC is also commonly used to validate other binding assays.
ITC is becoming an important tool in characterizing drug-target interactions, and can be used in different stages of Drug Discovery and Development. ITC is versatile and can be used to characterize binding between any two molecules.
Enzyme Kinetics
Enzyme catalytic reactions are central to all biological pathways. A major portion of biochemical research is devoted to characterizing enzyme function, activity and structure, and how enzymes are inhibited and activated. Enzyme characterization is also key to drug discovery since many drug targets are enzymes. These include kinases and proteases.
Isothermal Titration Calorimetry (ITC) is well-established in the study of affinity of molecular interactions, and is now becoming a mainstream tool in the study of enzyme kinetics. The strength of the technique lies in the universal nature of ITC. Traditional enzyme assays utilize a probe to monitor either substrate depletion or product formation. These probes are system-dependent and must be optimized for each reaction under specific conditions. Also, the substrate may need to be modified which could interfere with the catalysis reaction. For optical methods, the experimental conditions can affect the detection system, preventing accurate measurements. This means that with traditional assay methods, many enzymes do not have practical assays.
ITC uses heat as a probe, and since every reaction generates or absorbs heat, there is no need for lengthy method development each time a new enzyme is assayed. ITC directly measures the heat change as catalysis proceeds, which is proportional to the rate of reaction. Todd and Gomez (2001) showed that Km and kcat from ITC experiments agreed favorably with traditional enzyme kinetics methods, and can be used with every class of enzyme, including those with no other direct assay methods.
The use of ITC to monitor the rate of enzymatic reactions is a non-destructive, sensitive, and direct assay. Multiple injections of substrate can be done in a single experiment, so Km and kcat can be determined in a single ITC experiment. With ITC, it is straightforward to vary experimental conditions such as pH and ionic strength, and one can get a complete analysis of catalysis and kinetics. ITC also provides valuable insights on the thermodynamics of enzymatic reactions.
Measurement of Tight Binding Affinities with ITC
The range of binding constants which can be directly measured with Isothermal Titration Calorimetry (ITC) range from millimolar to nanomolar. With tight binding (KB >109 M-1 or Kd tighter than nanomolar), ITC titrations lose their curvature and affinity cannot be accurately calculated (Figure1, panel A – Wiseman et al, 1989). A method to measure KB greater than 109 M-1 has been developed (Sigurskjold, 2000, Velazquez-Campoy and Freire, 2006). This method requires two ligands : a tight binding ligand (A) and a second ligand (B) which has a weaker binding affinity and binds competitively to the same site as ligand A. In the first ITC experiment (Figure 1, panel B), ligand B is titrated into the protein solution, and KB and ΔH are determined. In the second ITC experiment (Figure 1, panel C), ligand A is titrated into the protein-ligand B complex and ligand A displaces ligand B. Using appropriate curve-fitting models (Sigurskjold, 2000) the binding constant of the tight-binding ligand can be calculated. Velazquez-Campoy et al (2001) have used this method to study several inhibitors of HIV-1 protease.
FIGURE 1. Representative competitive ligand binding ITC titrations for determination of KB for tight binding ligand.



The temperature dependence of binding heats is a phenomenon often overlooked by users of ITC. Consequently, experimental studies may be abandoned when investigators measure the binding enthalpy of a reaction at a temperature where the heat of binding is near zero. In such a case, the signal to noise ratio is poor, the binding isotherm is not well defined, and the heats of dilution are comparable to the reaction heats. The investigator may conclude that ITC is not suitable to study the interaction of interest. On the contrary, the temperature dependence of the heat of reaction is a valuable tool for optimizing the study of any binding event.
The heat of binding for a given reaction and the enthalpy change (ΔH) is typically temperature dependent. As you change the temperature of an experiment, the raw injection heat and therefore ΔH, change as well. The temperature dependence is due to the heat capacity change of the event (ΔCp = (ΔH1-ΔH2)/(T1-T2)). The value of ΔCp for biological interactions is almost always negative and ranges from approximately 0.3 to 2 kcal/degree/mole. If you collect a series of experiments using the same binding partners at different temperatures and plot the fitted values of ΔH vs. temperature, then the slope of a straight line through the data is the ΔCp. The ΔCp may be used to obtain higher heats of reaction and therefore better data. For example, suppose that you are currently working at 30°C and have obtained a fitted value for ΔH = -4 kcal/mol. Because binding enthalpy is temperature dependent, every degree that you increase the temperature of an experiment will increase the binding enthalpy 0.3 to 2 kcal/mole. The raw heats will increase as well. In this case, if you increase your experimental temperature to 37°C then you should obtain a binding enthalpy between -6.4 and -18 kcal/mole, higher raw heats, greater signal to noise, and a better defined binding curve. This is accomplished simply by changing the temperature of your experiment. You do not need to change the concentrations of your reactants. Alternatively, you could conduct the experiment at a lower temperature. Consider the above example. If the same experiment were performed at 5°C, then the binding enthalpy and raw heats will become more endothermic. If the ΔCp is -1 kcal/deg/mol, then reducing the experimental temperature to 5 degrees will yield a binding enthalpy of +21 kcal/mol. The raw injection heats will be endothermic as well and of greater magnitude than the original exothermic heats observed at 30°C. Since the ΔCp is usually linear and negative, then a measurement of binding enthalpy at two different temperatures will allow prediction of binding enthalpy at any temperature by fitting the data to a straight line.
Heat capacity change of binding is not a new concept. Long time users of MicroCal instruments have used the temperature dependence of binding heats to optimize experiments and obtain additional structural information associated with binding reactions. Most binding reactions have a temperature at which ΔH = 0. If you collect data within 5 to 10 degrees of this temperature, then the heats are almost always going to be low. If you change the temperature of the experiment, then you will increase the heat of the reaction and obtain better signal to noise. Interestingly, many systems have values for ΔH that go through zero between 20 and 30 degrees.
Accueil Présentation Recherche Formation Plateformes technologiques Médiation scientifique Stras-RNA Salon CNRS [Nom tutelle large] [Nom tutelle large] Rechercher Annuaire Intranet lien externe Accès restreint Accueil > Recherche > Équipes & Axes de Recherche > Structure et dynamique des machines biomoléculaires – E. Ennifar > ITC microcalorimetry ITC microcalorimetry Contact : Eric Ennifar Documentation ITC Data analysis in Origin ® – Tutorial guide ITC derived binding constants ITC200 manual Some interesting reviews and papers about ITC The use of biophysical methods increases success in obtaining liganded crystal structures Isothermal titration calorimetry of RNA Applications of Isothermal Titration Calorimetry in RNA Biochemistry and Biophysics Heat Capacity Changes Associated with Nucleic Acid Folding Calorimetry and Thermodynamics in Drug Design Ligand binding to one-dimensional lattice-like macromolecules : Analysis of the McGhee–von Hippel theory implemented in isothermal titration calorimetry ITC in the post-genomic era. . .? Priceless Isothermal titration calorimetry to determine association constants for high-affinity ligands Thermodynamics of Aminogl.–rRNA Recognition Useful Links Microcal TA Instruments Grants and Acknowledgments See paper : kinITC : a new method for obtaining joint thermodynamic and kinetic data by Isothermal Titration Calorimetry D. Burnouf, E. Ennifar, S. Guedich, B. Puffer, G. Hoffmann, G. Bec, F. Disdier, M. Baltzinger & P. Dumas JACS (2011) DOI : 10.1021/ja209057d http://pubs.acs.org/doi/abs/10.1021/ja209057d Supplementary Information Isothermal Titration Calorimetry (ITC) (microcal website) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method. What do you need for an experiment ? Contact me to take an appointment E.Ennifar@ibmc-cnrs.unistra.fr or K.Brillet@ibmc-cnrs.unistra.fr and to book a time slot. We ask 168.77 € HT for CNRS and Strasbourg University labs or 383.56 € HT for external labs for each 4 h time slot. A typical ITC experiment requires than 1 hour. ITC200 is the most sensitive isothermal titration calorimeter available, but it is also extremely fragile. For that reason, I will take care of it and perform your experiment(s) (but not process your data !) Filter or centrifuge your samples (very important especially to avoid any obstruction of the syringe) Samples requirement : 300 µl of sample in the cell 70 µl of ligand in the syringe (up to 200 µl if possible) 10 ml of buffer solution Sample and ligand have to be strictly in the same buffer (salts, pH …). This is extremely important for the ITC measurement ! Any difference will generate noise and make interpretation more difficult. To approximate the concentration needed in the cell, estimate the Kd for the system and multiply by 10. With very tight binders, the concentration should be on the low side of this value ; for weak binders, the concentration should be on the high side of this value. Multiply the cell concentration by n and by 10 – 15 for the syringe concentration. The minimum and maximum concentrations that are commonly used in the ITC200 are 3 µM to 500 µM. ΔS and ΔG are deduced from observed Kd and ΔH : ΔG = -RT log (1/Kd) = ΔH – T ΔS The binding reaction should be either endothermic or exothermic (ΔH ≠ 0), otherwise no signal will be observed An optimal determination of the Kd is obtained if 1000 > c > 1, where c = number of sites x Ka x concentration. For an optimal determination of the ΔH, a plateau should be obtained at the beginning of the experiment Process your data either with your favourite program or with Origin 7 installed on the ITC200 computer (see the manual below) Performance specifications : Operating temperature range : 2 – 80°C Titration sensitivity quotient : 8 ncal × ml Response time : 10 s Cell : 200 µl Injection syringe : 40 µl Smallest injection : 0.1 µl TC experiment performed on the Microcal ITC200 : successive injections of 1.4 µl lividomycin aminoglycoside 250 µM in 12 µM HIV-1 DIS RNA Single Injection Method (SIM) The ITC200 is now capable of carrying out a complete binding experiment using only a single continuous injection as opposed to the normal procedure which requires multiple injections. In this single-injection procedure, only one slow, continuous injection of ligand solution is made from the injection syringe into the macromolecule solution contained in the sample cell. The SIM technique significantly improves the speed of the experiment (typically 6 – 12 minutes per run !) and is particularly appropriate for drug discovery and target screening. Data analysis should be performed with the Origin Microcal LLC SIM module. Example of SIM performed on our ITC200 (19/01/2009) and comparison with an equivalent multiple injection run : Multiple 1.4 µl injections of 250 µM lividomycin aminoglycoside into a 12 µM HIV-1 DIS RNA solution. Single injection of 36 µl lividomycin 250 µM into a 12 µM HIV-1 DIS RNA solution. More information about the SIM is available on the Microcal website (http://www.microcal.com/products/itc/single-injection-method.asp). Also have a look into documentations available on this site. What is ITC ? (from Microcal website) Isothermal Titration Calorimetry (ITC) is the gold standard for measuring biomolecular interactions. ITC simultaneously determines all binding parameters (n, K, ΔH and ΔS) in a single experiment – information that cannot be obtained from any other method. When substances bind, heat is either generated or absorbed. ITC is a thermodynamic technique that directly measures the heat released or absorbed during a biomolecular binding event. Measurement of this heat allows accurate determination of binding constants (KB), reaction stoichiometry (n), enthalpy (ΔH) and entropy (ΔS), thereby providing a complete thermodynamic profile of the molecular interaction in a single experiment. Because ITC goes beyond binding affinities and can elucidate the mechanism of the molecular interaction, it has become the method of choice for characterizing biomolecular interactions. Applications include : Characterization of molecular interactions of small molecules, proteins, antibodies, nucleic acids, lipids and other biomolecules. Lead optimization. Enzyme kinetics. Assessment of the effect of molecular structure changes on binding mechanisms. Assessment of biological activitiy. Binding Studies All cellular processes require specific binding and molecular recognition between biomolecules. Knowledge of these interactions is important to understand how proteins, nucleic acids and lipids function in biological systems. There have been rapid advances in structural biology and relating structure to biochemical function and mechanism. However, knowledge of structure alone does not ensure accurate prediction of function and biological activity. The complete characterization of any binding interaction requires a quantification of the affinity, number of binding sites and the thermodynamics. Thermodynamic data, specifically enthalpy (ΔH) and entropy (ΔS), reveal the forces that drive complex formation and mechanism of action. Thermodynamics provide information on conformational changes, hydrogen bonding, hydrophobic interactions and charge-charge interactions. This information can be used to describe the function and mechanism at a molecular level. Binding Studies with Isothermal Titration Calorimetry (ITC) Isothermal Titration Calorimetry (ITC) is a powerful analytical tool which measures the binding affinity and thermodynamics between any two biomolecules. ITC is considered the “gold standard” assay for binding, and has many advantages : Universal assay – directly measures heat change associated with binding Label-free – uses native materials True in-solution technique Requires minimal assay development Has no molecular weight limitations Non-optical Versatile, can be used with any biomolecule – proteins, nucleic acids, small molecule drugs, lipids, etc. Can be used with a wide range of biological buffers, ionic strengths, pH’s In a single ITC experiment, one can determine : Binding affinity – Kd Directly measure sub-millimolar to nanomolar Can extend affinity range to picomolar using competitive ITC method Number of binding sites Multiple and different binding sites Enthalpy (ΔH) and entropy (ΔS) of binding ITC is vital in the study of multi-probe structure activity relationships (SAR) since it can detect contributions that affinity-only methods may miss. For example, the affinity measured by these methods may be similar for a wild-type and mutant protein binding to a drug, but ITC can reveal differences in ΔH and ΔS that can describe the mechanism of action of binding. This information can validate in-silico modeling. ITC is also commonly used to validate other binding assays. ITC is becoming an important tool in characterizing drug-target interactions, and can be used in different stages of Drug Discovery and Development. ITC is versatile and can be used to characterize binding between any two molecules. Enzyme Kinetics Enzyme catalytic reactions are central to all biological pathways. A major portion of biochemical research is devoted to characterizing enzyme function, activity and structure, and how enzymes are inhibited and activated. Enzyme characterization is also key to drug discovery since many drug targets are enzymes. These include kinases and proteases. Isothermal Titration Calorimetry (ITC) is well-established in the study of affinity of molecular interactions, and is now becoming a mainstream tool in the study of enzyme kinetics. The strength of the technique lies in the universal nature of ITC. Traditional enzyme assays utilize a probe to monitor either substrate depletion or product formation. These probes are system-dependent and must be optimized for each reaction under specific conditions. Also, the substrate may need to be modified which could interfere with the catalysis reaction. For optical methods, the experimental conditions can affect the detection system, preventing accurate measurements. This means that with traditional assay methods, many enzymes do not have practical assays. ITC uses heat as a probe, and since every reaction generates or absorbs heat, there is no need for lengthy method development each time a new enzyme is assayed. ITC directly measures the heat change as catalysis proceeds, which is proportional to the rate of reaction. Todd and Gomez (2001) showed that Km and kcat from ITC experiments agreed favorably with traditional enzyme kinetics methods, and can be used with every class of enzyme, including those with no other direct assay methods. The use of ITC to monitor the rate of enzymatic reactions is a non-destructive, sensitive, and direct assay. Multiple injections of substrate can be done in a single experiment, so Km and kcat can be determined in a single ITC experiment. With ITC, it is straightforward to vary experimental conditions such as pH and ionic strength, and one can get a complete analysis of catalysis and kinetics. ITC also provides valuable insights on the thermodynamics of enzymatic reactions. Measurement of Tight Binding Affinities with ITC The range of binding constants which can be directly measured with Isothermal Titration Calorimetry (ITC) range from millimolar to nanomolar. With tight binding (KB >109 M-1 or Kd tighter than nanomolar), ITC titrations lose their curvature and affinity cannot be accurately calculated (Figure1, panel A – Wiseman et al, 1989). A method to measure KB greater than 109 M-1 has been developed (Sigurskjold, 2000, Velazquez-Campoy and Freire, 2006). This method requires two ligands : a tight binding ligand (A) and a second ligand (B) which has a weaker binding affinity and binds competitively to the same site as ligand A. In the first ITC experiment (Figure 1, panel B), ligand B is titrated into the protein solution, and KB and ΔH are determined. In the second ITC experiment (Figure 1, panel C), ligand A is titrated into the protein-ligand B complex and ligand A displaces ligand B. Using appropriate curve-fitting models (Sigurskjold, 2000) the binding constant of the tight-binding ligand can be calculated. Velazquez-Campoy et al (2001) have used this method to study several inhibitors of HIV-1 protease. FIGURE 1. Representative competitive ligand binding ITC titrations for determination of KB for tight binding ligand. Panel A : Macromolecule in ITC cell, tight binding ligand A in syringe. Titration is not sigmoidal curve, so unable to determine KB for ligand A. Panel B : Macromolecule in ITC cell, weak binding ligand B in syringe Panel C Macromolecule plus ligand B in ITC cell, ligand A in syringe. During titration, ligand A binds to macromolecule, displacing ligand B. Using model of Sigurskjold, KB for ligand A is 1.2 x 109 M-1 Temperature dependence of binding heats The temperature dependence of binding heats is a phenomenon often overlooked by users of ITC. Consequently, experimental studies may be abandoned when investigators measure the binding enthalpy of a reaction at a temperature where the heat of binding is near zero. In such a case, the signal to noise ratio is poor, the binding isotherm is not well defined, and the heats of dilution are comparable to the reaction heats. The investigator may conclude that ITC is not suitable to study the interaction of interest. On the contrary, the temperature dependence of the heat of reaction is a valuable tool for optimizing the study of any binding event. The heat of binding for a given reaction and the enthalpy change (ΔH) is typically temperature dependent. As you change the temperature of an experiment, the raw injection heat and therefore ΔH, change as well. The temperature dependence is due to the heat capacity change of the event (ΔCp = (ΔH1-ΔH2)/(T1-T2)). The value of ΔCp for biological interactions is almost always negative and ranges from approximately 0.3 to 2 kcal/degree/mole. If you collect a series of experiments using the same binding partners at different temperatures and plot the fitted values of ΔH vs. temperature, then the slope of a straight line through the data is the ΔCp. The ΔCp may be used to obtain higher heats of reaction and therefore better data. For example, suppose that you are currently working at 30°C and have obtained a fitted value for ΔH = -4 kcal/mol. Because binding enthalpy is temperature dependent, every degree that you increase the temperature of an experiment will increase the binding enthalpy 0.3 to 2 kcal/mole. The raw heats will increase as well. In this case, if you increase your experimental temperature to 37°C then you should obtain a binding enthalpy between -6.4 and -18 kcal/mole, higher raw heats, greater signal to noise, and a better defined binding curve. This is accomplished simply by changing the temperature of your experiment. You do not need to change the concentrations of your reactants. Alternatively, you could conduct the experiment at a lower temperature. Consider the above example. If the same experiment were performed at 5°C, then the binding enthalpy and raw heats will become more endothermic. If the ΔCp is -1 kcal/deg/mol, then reducing the experimental temperature to 5 degrees will yield a binding enthalpy of +21 kcal/mol. The raw injection heats will be endothermic as well and of greater magnitude than the original exothermic heats observed at 30°C. Since the ΔCp is usually linear and negative, then a measurement of binding enthalpy at two different temperatures will allow prediction of binding enthalpy at any temperature by fitting the data to a straight line. Heat capacity change of binding is not a new concept. Long time users of MicroCal instruments have used the temperature dependence of binding heats to optimize experiments and obtain additional structural information associated with binding reactions. Most binding reactions have a temperature at which ΔH = 0. If you collect data within 5 to 10 degrees of this temperature, then the heats are almost always going to be low. If you change the temperature of the experiment, then you will increase the heat of the reaction and obtain better signal to noise. Interestingly, many systems have values for ΔH that go through zero between 20 and 30 degrees. Why “N” might not turn out as you expect… “N” is the average number of binding sites per mole of protein in your solution, assuming:
- all binding sites are identical and independent
- you have pure protein (and ligand)
- you have given the correct protein and ligand concentrations
- all your protein is correctly folded and active…
This is rarely true in practice ! Protein (and ligand) concentration determinations depend on the accuracy of the methods used. Protein extinction coefficients, for example, are rarely known better than ±5%, and are usually worse. Poor measurement techniques, incorrect UV baseline corrections, attempts to conserve material using “micro” cuvettes for example can lead to serious errors. Even if all your measurements are dead accurate, not all the protein may be correctly folded (a common experience with recombinant proteins).
Possible cases:
N
- protein concentration is lower than you think, or…
- protein is impure, or…
- protein (polypeptide) is pure but not all correctly folded, or…
- ligand concentration is higher than you think, or…
- simple non-cooperative binding model is inappropriate, or…
- all of the above
N > 1
- your protein has multiple binding sites, or…
- ligand concentration is lower than you think, or…
- simple non ?cooperative binding model is inappropriate, or…
- all of the above
No measurable signal, if ΔH close to 0 …
Kd out of range for an ITC experiment : if c 1000 displacement of a competitor might be required.
No useful information can be obtained from an unspecific binding…
Other problems:

 Intranet
Intranet Access
Access Contact
Contact



