Our goal is to understand the molecular mechanism of biomolecular machineries using an integrated biophysics approach, combining biochemistry, thermodynamics, kinetics, structural biology and molecular dynamics.

Research axis
Contacts: Eric Ennifar, Isabelle Lebars.
Collaborateurs: Joao Marques (Federal University of Minas Gerais, Brazil & CNRS UPR 9022 IBMC), Karim Majzoub (IGMM Montpellier, CNRS UMR 5535).
Zika (ZIKV) and Dengue (DENV) are mosquito-borne viruses belonging to the Flaviviridae family. With an estimated 390 million new infections every year, DENV is considered by the World Health Organization to be the most critical mosquito-borne viral disease worldwide. Regarding ZIKV, it was a relatively obscure pathogen until 2007, when it started causing outbreaks in Pacific islands and later in the Americas, with a massive outbreak in Brazil in 2015-2016. Currently, no efficient treatments or vaccines are available against these viruses. Considering that DENV and ZIKV circulate in both vertebrate and invertebrate hosts, targeting specific steps of viral replication would be an efficient strategy to block viral transmission and disease spread. The development of new specific antiviral strategies against ZIKV and DENV requires a precise understanding of key steps of the viral lifecycle. RNA viruses often harbor, within their genomes, multifunctional non-coding structured regions. The 5’ and 3’ untranslated regions (UTR) in the genomes of DENV and ZIKV are very conserved and have important functions during viral replication.

RNA-protein interactions play a crucial role in the viral replication cycle and for hijacking host cell machinery. In order to develop new antiviral strategies, our aims are to : (i) decipher RNA-protein interactions involved in DENV and ZIKV replication cycle and (ii) identify and characterize mosquito factors interacting with the RNA genome of these viruses. For that purpose, we use an integrated biophysical approach including Isothermal microcalorimetry (ITC), electro switchable DNA nanolevers on biochips using the switchSENSE biosensor technology, UV-visible spectrometry, Mass Spectrometry (MS), Electron Paramagnetic Resonance (EPR, in collaboration), CryoElectron Microscopy (CryoEM), Nuclear Magnetic Resonance (NMR) and X-ray Crystallography.
coming soon
In eukaryotic cells, translation initiation begins with the recruitment of the 43S pre-initiation complex (43S PCI) composed of the small ribosomal subunit (40S) and several initiation factors (eIFs) to the 5′ extremity of an mRNA using a m7G ‘cap’ structure, facilitated by the cap-binding complex (eIF4F). Subsequently, the eIFs bound to the 43S PIC assist in the reliable recognition of the mRNA start codon, leading to the formation of the late-stage 48S IC (LS48S IC). Upon the joining of the large ribosomal subunit (60S), the 80S IC is formed. Although predominant for most cellular mRNAs, this classical pattern is compromised under various stress conditions, especially those related to the tumor cell microenvironment, such as hypoxia, nutrient starvation, and immune response. Solid tumor cells are often subjected to chronic hypoxia, resulting in a cyclic transition from a hypoxic to a normoxic state which induced a translation reprogramming. However, the translation of a subset of cellular mRNA coding for regulators of tumor development is maintained by alternative mechanisms involving RNA-binding proteins or elements contained within the mRNA itself (IRES – Internal Ribosome Entry Sites, RNA modifications, RNA G-quadruplexes, µORFs), often found in the 5’UTR of their mRNA. Among them, mRNA for hypoxia-inducible factor 1α (HIF-1α) and β-catenin are particularly interesting because the maintenance of their translation promotes the persistence and plasticity of cancer cells. Currently, the existence of an alternative translation mechanism supporting HIF-1α and β-catenin mRNA translation under hypoxia has not been confirmed, and several questions remain open: i) what are the structural and/or sequence determinants for their translation when canonical initiation is altered? ii) How does the ribosome interact with these determinants? Is the m7G ‘cap’ structure required? Are there specific eIFs and/or trans-acting factors required?

The main objective of our research is to gain atomic-level insight into the mechanism of hypoxia-induced alternative translation initiation regulating HIF-1α and β-catenin expression using a combination of biochemical (RNA structure mapping, in vitro translation, and cellular RNA transfection), biophysical (thermodynamics and kinetics), and structural biology (X-ray crystallography and cryo-electron microscopy) approaches.
Thus, this study should identify structural features of mRNAs and/or factors playing a central role in HIF-1α and β-catenin mRNA translation during O2 deprivation, which may become targets for a new class of anticancer drugs.

coming soon
coming soon

Past Projects
Contact: Isabelle LEBARS.
Collaborator: Dr Anne-Catherine Dock-Bregeon (LBI2M, Roscoff, CNRS UMR 8227).
The 7SK small nuclear RNA (7SKsnRNA) plays a key role in the regulation of RNA polymerase II by sequestrating and inhibiting the positive transcription elongation factor b (P-TEFb) in the 7SK ribonucleoprotein complex (7SKsnRNP), a process mediated by interaction with the protein HEXIM. P-TEFb, a heterodimer comprising the cyclin T1 and the cyclin-dependent kinase CDK9, is also an essential cellular factor recruited by the viral protein Tat to ensure the replication of the viral RNA in the infection cycle of the human immunodeficiency virus (HIV-1). Tat promotes the release of P-TEFb from the 7SKsnRNP and subsequent activation of transcription, by displacing HEXIM from the 5’-hairpin of the 7SKsnRNA.

This hairpin (HP1), comprising the signature sequence of the 7SKsnRNA, has been the subject of three independent structural studies aiming at identifying the structural features that could drive the recognition by the two proteins (HEXIM and Tat), both depending on similar Arginine Rich Motifs (ARM). Interestingly, four distinct structures were determined under different conditions (pH, salts). We used Isothermal Titration Calorimetry (ITC) to determine the thermodynamic signatures of the Tat-ARM and HEXIM-ARM peptide interactions with the RNA under different conditions, and we show that they are associated with distinct binding mechanisms.
Publication:
Brillet K, Martinez-Zapien D, Bec G, Ennifar E, Dock-Bregeon A C, Lebars I. Different views of the dynamic landscape covered by the 5′-hairpin of the 7SK small nuclear RNA. RNA (2020), 26 (9):1184-1197.https://pubmed.ncbi.nlm.nih.gov/32430362/
Financial support:

HIV-1 reverse transcriptase (RT) is a key enzyme in the life cycle of the virus, converting viral genomic RNA into proviral DNA. It is also a cornerstone of anti-HIV therapy, as half of the individual compounds that inhibit viral replication target the RT polymerase active site. HIV-1 RT inhibitors are divided into nucleoside-like RT inhibitors (NRTIs), which compete with the natural nucleoside substrate, and non-nucleoside RT inhibitors (NNRTIs), which bind to a hydrophobic pocket adjacent to the polymerase active site. We performed biophysical and structural studies on two new RT inhibitors: a DABO analog that emerged as one of the most active RT inhibitors reported to date (Ec50 = 25 pM), and DAVP-1, a non-nucleotide RT inhibitor with an unusual mechanism of action. The crystal structure of HIV-1 RT bound to DAVP-1 revealed a novel binding site close to the catalytic site of RT polymerase.
We also addressed the mechanism of action of RT and non-nucleoside RT inhibitors (NNRTIs) by isothermal titration calorimetry (ITC). Using a novel incremental ITC approach, a step-by-step thermodynamic dissection of RT polymerization activity showed that most of the driving force for DNA synthesis is provided by initial dNTP binding. Surprisingly, the thermodynamic and kinetic data led to a reinterpretation of the NNRTI inhibition mechanism. NNRTI binding to preformed RT/DNA complexes is hindered by a kinetic barrier and NNRTIs interact primarily with free RT. Once formed, RT/INNTI complexes either bind to DNA in an apparent polymerase-compatible orientation or form high-affinity dead-end complexes, with both RT/INNTI/DNA complexes unable to bind the incoming nucleotide substrate.



Selected publications:
- Bec G, Meyer B, Gerard MA, Steger J, Fauster K, Wolff P, Burnouf D, Micura R, Dumas P & Ennifar E (2013). Thermodynamics of HIV-1 Reverse Transcriptase in action elucidates the mechanism of action for non-nucleoside inhibitors. J Am Chem Soc, 135(26):9743-52.
- Freisz S, Bec G, Radi M, Wolff P, Crespan E, Angeli L, Dumas P, Maga G, Botta M & Ennifar E (2010). Crystal Structure of the HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action. Angew Chem Int Ed Engl, 49(10):180
Riboswitches are non-coding elements upstream or downstream of messenger RNAs that, by binding to a specific ligand, regulate transcription or translation initiation in bacteria, or alternative splicing in plants and fungi. We studied the aptamer domains of the thiamine pyrophosphate riboswitches (TPP) of thiC and thiM in E. coli, which regulate transcription and translation, respectively, and that of THIC in A. thaliana. For all of them, we established an induced adjustment mechanism with an initial loose binding of TPP (characterized by the kon and koff parameters) subsequently transformed into a tight binding after an RNA conformational change (characterized by the kF and kU parameters). Using our kinITC approach, we obtained the temperature dependence of all kinetic and thermodynamic parameters for bacterial riboswitches. The results imply kinetic regulation, which requires RNA polymerase to stop after the synthesis of the riboswitch aptamer. A quantitative model of regulation highlighted how the pause time must be related to the initial TPP binding kinetic rates to achieve an ON/OFF switch in the correct TPP concentration range. We verified the existence of these pauses and the model’s prediction of their duration. The A. thaliana riboswitch, in contrast, is under classical thermodynamic control and showed considerable temperature dependence of its folding kinetics, suggesting that it may also act as a thermosensor. Our analysis also showed that kinetically regulated riboswitches respond more strongly to changes in ligand concentration than thermodynamically regulated riboswitches. This rationalizes the interest of kinetic regulation

Selected publications:
- Guedich S., Puffer-Enders B., Baltzinger M., Hoffmann G., Da Veiga C., Jossinet F., Thore S., Bec G., Ennifar E., Burnouf D., Dumas P. (2016). Quantitative and predictive model of kinetic regulation by E. coli TPP riboswitches. RNA Biol, 13(4):373-90.
- Burnouf D.Y., Ennifar E., Guedich S., Puffer B., Hoffmann G., Bec G., Disdier F., Baltzinger M, Dumas P. (2012). kinITC : a new method for obtaining joint thermodynamic and kinetic data by Isothermal Titration Calorimetry. J Am Chem Soc, 134(1):559-565.
The HIV-1 dimerization initiation site (DIS) is a conserved hairpin in the 5′ UTR of genomic RNA. Alteration of DIS affects RNA dimerization, packaging, and reverse transcription, and reduces viral infectivity. The DIS loop initiates genome dimerization by forming a loop-loop complex and is subsequently stabilized, presumably in an extended duplex form by interaction with the viral NCp7 nucleocapsid protein. We solved the crystal structures of the DIS kissing loop and the extended duplex. Our structures revealed similarities to the A-site of the bacterial 16S ribosomal RNA, which is the target of aminoglycoside antibiotics. As a result, we showed that aminoglycosides bind to HIV-1 DIS. Surprisingly, the affinity of these molecules for the kissing loop of DIS is higher than for their natural target in bacterial ribosomes. Their binding strongly stabilizes the loop-loop complex and prevents its conversion to duplex form, even in the presence of NCp7. We have also solved numerous high-resolution X-ray structures of the DIS kissing-loop complex and the extended duplex bound to several aminoglycosides. However, identification of the molecular driving forces important for DIS/aminoglycoside binding could not be achieved by considering structural data alone, highlighting the importance of collecting thermodynamic and structural data for a complete understanding of the molecular recognition process.
Using ITC microcalorimetry, the thermodynamics of aminoglycosides binding to the duplex and kissing-loop forms of HIV-1 DIS was studied under a range of conditions. With the structural data provided by high-resolution crystal structures, we were able to establish the basis for the specificity of drug/RNA recognition and allow discrimination between specific binding sites and potential competing secondary sites. We were also able to show that drug binding to the DIS kissing-loop complex inhibits NCp7-assisted conversion in the extended duplex form. Using all these data, we could rationally design an aminoglycoside conjugate that specifically binds to HIV-1 DIS RNA. Our results show that it is possible to target the HIV-1 DIS dimer before and after NCp7-assisted RNA maturation with the same molecule.


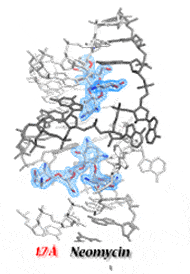
Selected publications:
- Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P (2001). Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol, 8(12):1064-8.
- Ennifar E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C, Dumas P, Walter P (2003). HIV-1 RNA dimerization initiation site is structurally similar to the ribosomal A site and binds aminoglycoside antibiotics. J Biol Chem, 278(4):2723-30.
- Bernacchi S, Ennifar E, Tóth K, Walter P, Langowski J, Dumas P (2005). Mechanism of hairpin-duplex conversion for the HIV-1 dimerization initiation site. J Biol Chem, 280(48):40112-21.
- Ennifar E, Paillart JC, Bodlenner A, Walter P, Weibel JM, Aubertin AM, Pale P, Dumas P, Marquet R (2006). Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides : from crystal to cell. Nucleic Acids Res, 34(8):2328-39.
- Bernacchi S, Freisz S, Maechling C, Spiess B, Marquet R, Dumas P, Ennifar E (2007).Aminoglycoside binding to the HIV-1 RNA dimerization initiation site : thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res, 35(21):7128-39.
- Freisz S, Lang K, Micura R, Dumas P, Ennifar E (2008). Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site. Angew Chem Int Ed Engl, 47(22):4110-3.
- Ennifar E, Aslam MW, Strasser P, Hoffmann G, Dumas P, van Delft FL (2013). Structure-guided discovery of a novel aminoglycoside conjugate targeting HIV-1 RNA viral genome. ACS Chem Biol, 8(11):2509-17.
to be completed
Selected publications:
- Freisz S., Mezher J., Hafirassou L., Wolff P., Nomine Y., Romier C., Dumas P., Ennifar E. (2012). Sequence and structure requirements for specific recognition of HIV-1 TAR and DIS RNA by the HIV-1 Vif protein. RNA Biol, 9(7):966 – 977.
We have solved a 27-nt RNA structure at subatomic resolution (0.57 Å). At this resolution, most of the hydrogen atoms in the RNA, as well as some hydrogens in the aqueous solvent, are visible. Furthermore, at such ultimate resolution, the model of independent spherical atoms cannot be applied and strain densities are visible. Therefore, we are currently refining the structure using multipole refinement methods.

Development of a new stepwise approach: the RIP method. We had reported that one of the main causes of failure of the MAD technique with bromo- or iodo-uridines was due to X-ray induced cleavage of the halogen atom. By considering the cleavage of these anomalous scatterers as generating a “native” from a “derived” crystal, we were able to complement the progressive loss of the anomalous signal used in the MAD phasing by this concomitant MIR-like signal. We have now strengthened the method by showing how to monitor the extent of cleavage at any time during data collection.
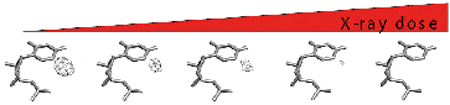
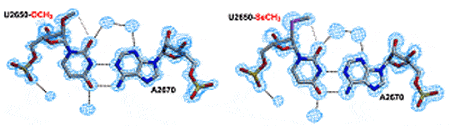
An alternative for MAD phasing is to use selenized RNA (increased radiation resistance). However, modification of RNA by replacing one or more 2′-OH groups with one or more 2′-SeCH3 groups is chemically cumbersome and expensive. Therefore, we developed a strategy based on preliminary screening of 2′-OCH3-modified RNA crystallization, prior to replacement of the 2′-OCH3 groups with their 2′-SeCH3 counterparts.

We have implemented major methodological developments in isothermal titration calorimetry (ITC). First, ITCs that provide kinetic information similar to data usually obtained by surface plasmon resonance, as well as thermodynamic information usually recovered by ITCs. In partnership with Software for Science Development, our kinITC strategy has been implemented in the AFFINIMETER ITC data processing software
Second, incremental ITC allows step-by-step dissection of successive reactions in biomolecular machines. This approach has been successfully used to discover the mechanism of action of HIV-1 reverse transcriptase and the mechanism of inhibition of HIV-1 reverse transcriptase inhibitors used in the clinic.

Multimeric sliding clamps for DNA (also known as β-rings) confer high processivity to replicative DNA polymerases. They are also molecular centers on which many proteins involved in DNA metabolism interact by binding, via a conserved peptide sequence, in a universally conserved pocket (Figure).
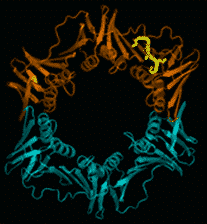
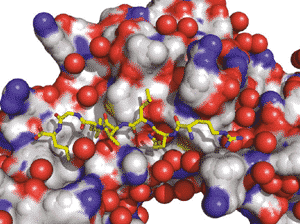
In bacteria, the interaction pocket is a potential new target for the development of new antibacterial compounds, which are urgently needed to control and overcome the growing bacterial resistance to antibiotics.
We use a structure-based approach to design peptides that bind to the interaction pocket with high affinity, compete with DNA polymerases and trigger cell death. This project is conducted through a multidisciplinary and integrated strategy resulting from a close collaboration between seven different laboratories, combining chemical synthesis, molecular modeling, structural, biophysical and biochemical analyses and in vivo evaluation of the ligands’ efficacy to inhibit bacterial growth and infectious processes.
In this collaboration, our task is to produce β-rings from different bacterial origins and to analyze the interaction of newly designed peptides with these rings, using biophysical techniques such as ITC, X-ray crystallography and mass spectrometry.
Selected publications:
- Burnouf D., Olieric V., Wagner J., Fujii S., Reinbolt J., Fuchs R.P. and Dumas P. (2004). Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J Mol Biol, 335(5):1187-1197.
- Wolff P., Olieric V., Briand J.P., Chaloin O., Dejaegere A., Dumas P., Ennifar E., Guichard G., Wagner J., Burnouf D. (2011). Structure-based design of short peptide ligands binding onto the E. coli processivity ring. J Med Chem, 54(13):4627-37.
- Wolff P., Amal I., Oliéric V., Chaloin O., Gygli G., Ennifar E., Lorber B., Guichard G., Wagner J., Dejaegere A. and Burnouf D. (2014). Differential modes of peptide binding onto replicative sliding clamps from various bacterial origins. J Med Chem., 57(18):7565-7576.
- Wolff P., Ennifar E., Guichard G., Burnouf D. and Dumas P. Native ESI mass spectroscopy can help avoiding wrong interpretations from isothermal titration calorimetry in difficult situations. Soumis à American Journal of Mass Spectrometry
Collaborations:
Prof Annick Dejaegere (IGBMC, Illkirch, France), Dr Gilles Guichard (IECB, Pessac, France), Dr Gaetan Mislin (ESBS, Illkirch, France), Dr Vincent Oliéric ( SLS, Villigen, Suisse), Prof Jean Marc Reichardt (IBMC, Strasbourg, France), Dr Jérôme Wagner (ESBS, Illkirch, France).
Financial supports:
Inserm, AstraZeneca
Patents:
- Structure cristalline de la protéine du facteur de processivité de l’ADN polymérase et un ligand et cet usage. EP 1639509, 29 mars 2003 – WO2004EP06942 20040625. Burnouf D, Wagner J, Dumas P, Fujii S, Fuchs R, Olieric V.
- Compounds binding to the bacterial beta ring. EP11162733.7, dépôt EPO le 15 avril 2011, délivré le 7 septembre 2012. Burnouf D, Dejaegere A, Guichard G, Oliéric V, Wagner J.
Selected publications
Simonetti A, Guca E, Bochler A, Kuhn L, Hashem Y. Structural Insights Into the Mammalian Late-Stage Initiation Complexes. Cell Rep (2020), 31 (1):107497. https://www.ncbi.nlm.nih.gov/pubmed/32268096?dopt=Abstract
Brillet K, Martinez-Zapien D, Bec G, Ennifar E, Dock-Bregeon A C, Lebars I. Different views of the dynamic landscape covered by the 5′-hairpin of the 7SK small nuclear RNA. RNA (2020), 26 (9):1184-1197.https://pubmed.ncbi.nlm.nih.gov/32430362/
Auffinger P, Ennifar E, D’Ascenzo L. Deflating the RNA Mg 2+ bubble. Stereochemistry to the rescue! RNA (2020), 27 (3):243-252. https://doi.org/10.1261/rna.076067.120
D’Ascenzo L, Vicens Q, Auffinger P. Identification of receptors for UNCG and GNRA Z-turns and their occurrence in rRNA. Nucleic Acids Res (2018), 46(15):7989-7997. https://www.ncbi.nlm.nih.gov/pubmed/29986118?dopt=Abstract
Bec G, Meyer B, Gerard MA, Steger J, Fauster K, Wolff P, Burnouf B, Micura R, Dumas P, Ennifar E. Thermodynamics of HIV-1 reverse transcriptase in action elucidates the mechanism of action of non-nucleoside inhibitors. J Am Chem Soc (2013);135(26):9743-52. https://pubmed.ncbi.nlm.nih.gov/23742167/
Ennifar E, Aslam MW, Strasser P, Hoffmann G, Dumas P, van Delft FL. Structure-guided discovery of a novel aminoglycoside conjugate targeting HIV-1 RNA viral genomeACS Chem Biol (2013);8(11):2509-17. https://pubmed.ncbi.nlm.nih.gov/24015986/
Burnouf D Y, Ennifar E, Guedich S, Puffer B, Hoffmann G, Bec G, Disdier F, Baltzinger M, Dumas P. kinITC: a new method for obtaining joint thermodynamic and kinetic data by Isothermal Titration Calorimetry. J Am Chem Soc (2012), 134(1):559-565.
http://www.ncbi.nlm.nih.gov/pubmed/22126339
Freisz S, Bec G, Radi M, Wolff P, Crespan E, Angeli L, Dumas P, Maga G, Botta M, Ennifar E. Crystal structure of HIV-1 reverse transcriptase bound to a non-nucleoside inhibitor with a novel mechanism of action. Angew Chem (2010), 49(10):1805-8. https://pubmed.ncbi.nlm.nih.gov/20135654/
Ennifar E, Paillart J C, Bodlenner A, Walter P, Weibel J M, Aubertin A M, Pale P, Dumas P, Marquet R. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell. Nucleic Acids Research (2006), 34(8):2328-39. https://pubmed.ncbi.nlm.nih.gov/16679451/
Financial supports



 Intranet
Intranet Access
Access Contact
Contact
