Selection of the HIV-1 genomic RNA and viral assembly
Like the genome of all retroviruses, the HIV-1 genomic RNA consists of two homologous RNA molecules of positive polarity. Dimerization and packaging are intricately linked processes. The Gag precursor (Pr55Gag in the case of HIV-1) plays a central role in the selection of the genomic RNA in the cytoplasm and in the assembly of the viral particles at the plasma membrane. The budding viral particles are immature and non infectious; they become infectious after maturation, a process induced by the cleavage of the Gag and GaPol precursors by the viral protease (which is part of GagPol).
- Selection of the genomic RNA relies on highly specific interaction with a small number of Pr55Gag molecules. Stem-loop 1 (SL1) whose apical loop mediates dimerization of the HIV-1 genomic RNA, plays a key role in Pr55Gag binding and a double regulation by the RNA regions flanking SL1 allows discrimination between the genomic RNA and spliced viral RNAs (Fig.1). Our goal is to identify the sequences and the interactions allowing this discrimination.
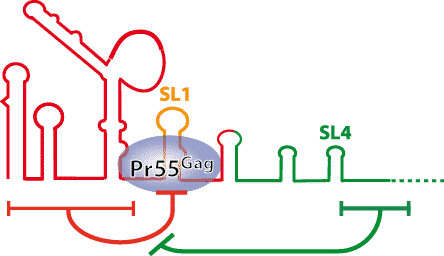
- During assembly of the immature viral particles, unspecific interactions between Pr55Gag and nucleic acids induce multimerization of 4,000 Gag precursor copies around the dimeric genomic RNA. So far, nobody knows if the change in specificity of Pr55Gag for RNA is a direct consequence of the accumulation of this protein in lipid rafts or if results from molecular signals (e.g. from a Pr55Gag conformational change resulting from the insertion of its N-terminal myristoyl moiety in the plasma membrane).
- During maturation of the viral particles, sequential cleavages of the Gag precursor generate the structural proteins of the mature virus. Maturation is accompanied by important structural rearrangements that are easily observed by electron microscopy (Fig. 2). Concomitantly, the genomic RNA dimer becomes more stable and more compact. The structural rearrangements of the genomic RNA remain largely unknown despite increasing evidence showing that there are required for the infectiveness of the mature viral particles.

Our goal is to elucidate the molecular mechanisms that govern selection of the genomic RNA and assembly of the viral particles by combining structural and functional studies, in vitro and in cell culture.
 Intranet
Intranet Access
Access Contact
Contact

